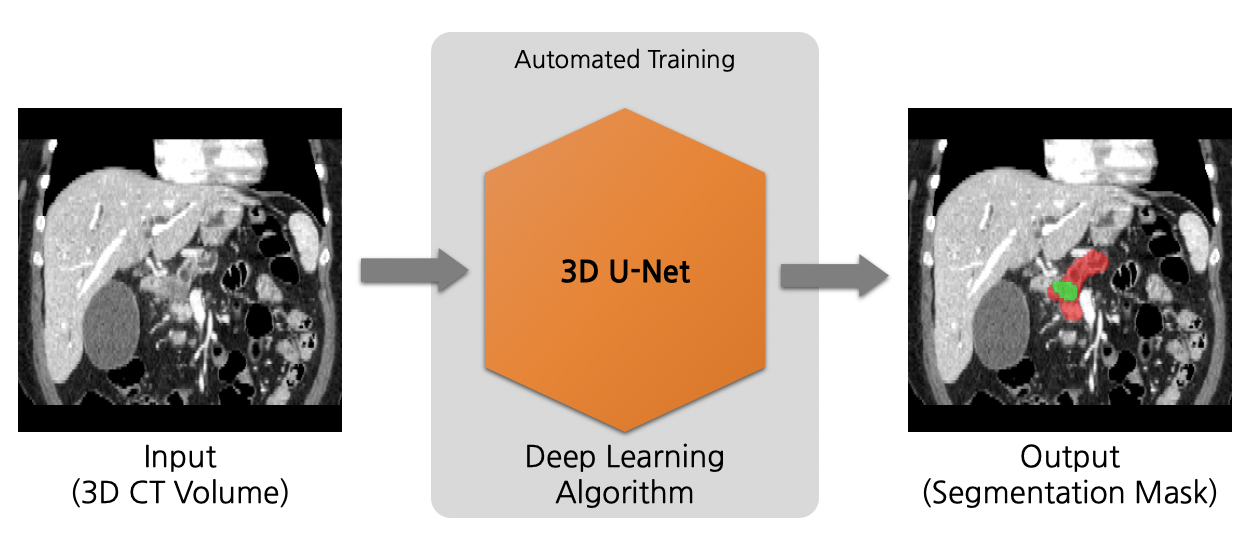
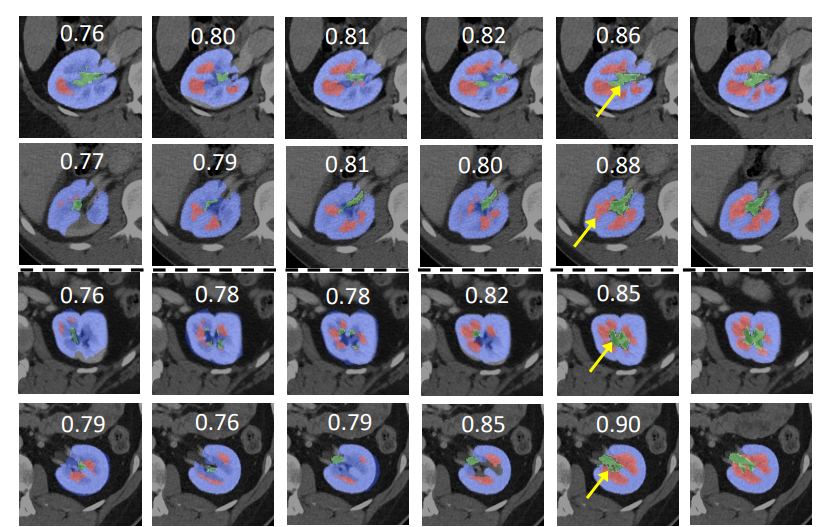
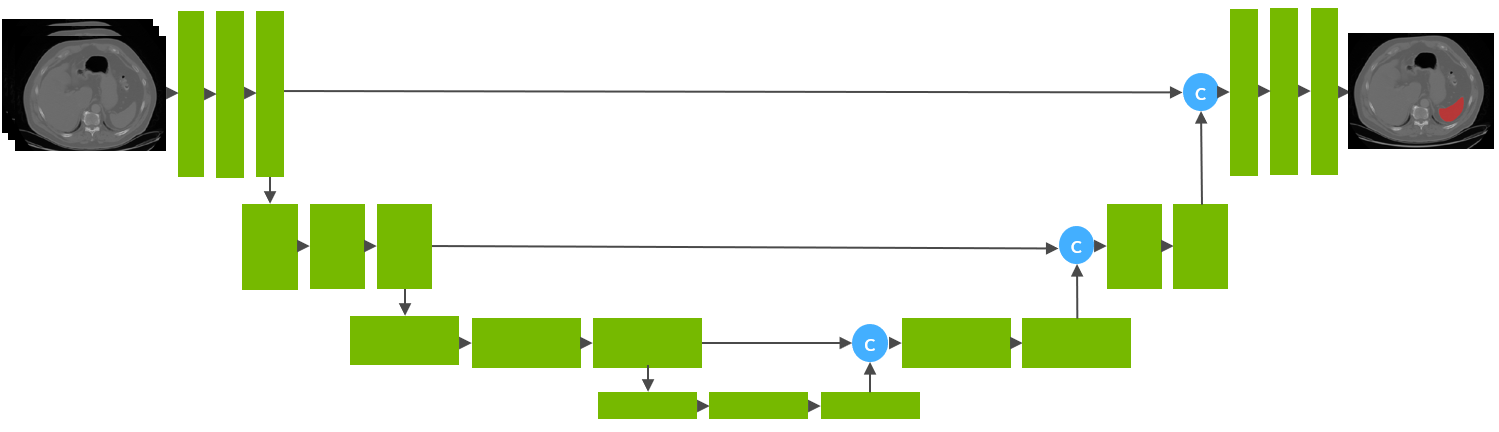
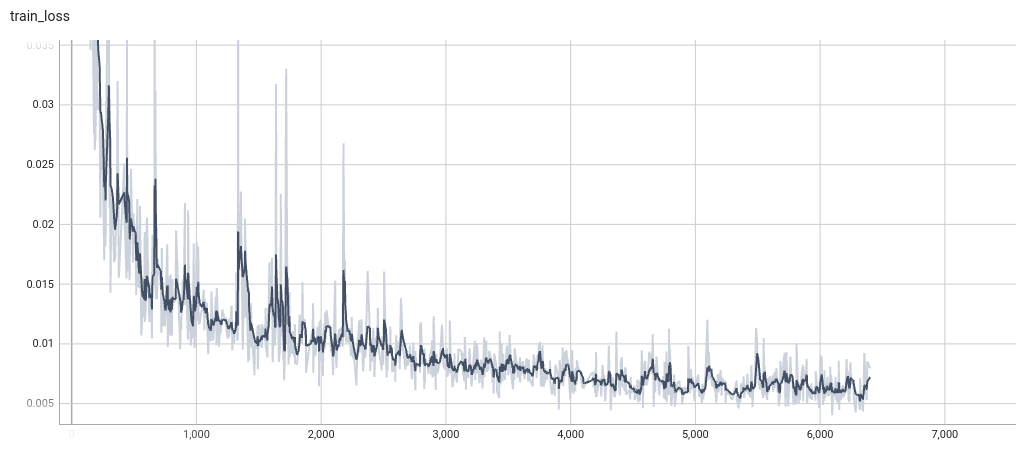
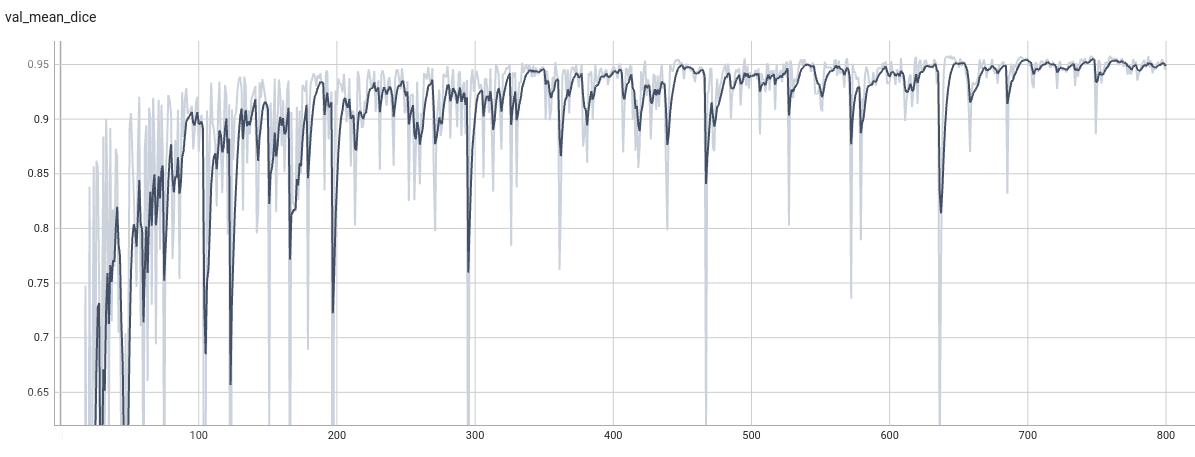
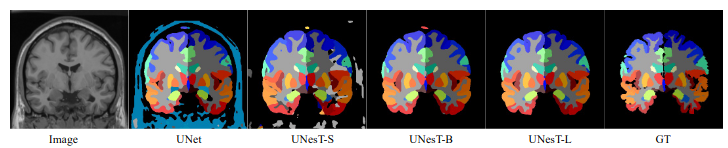
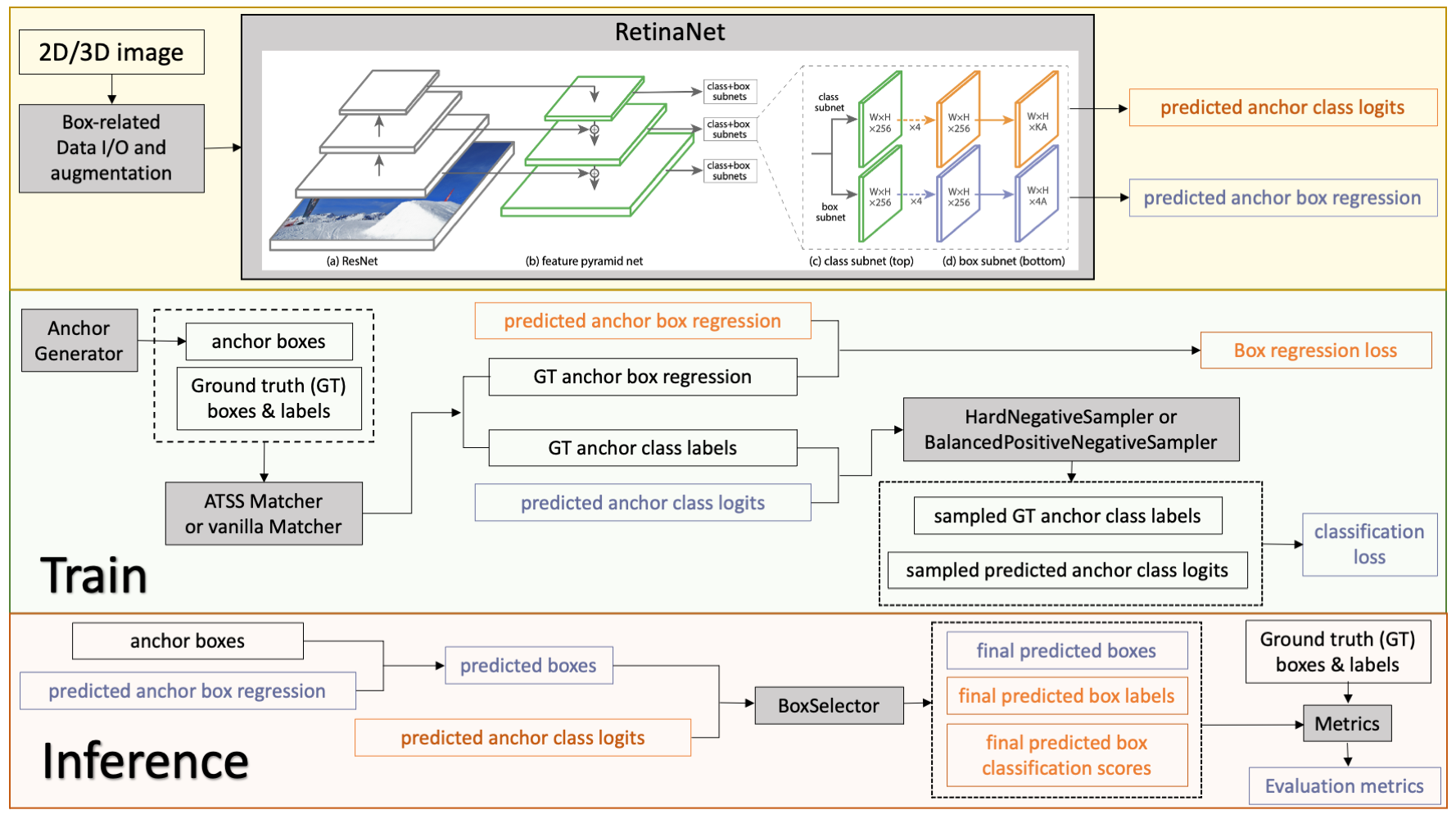
Model Overview
A pre-trained model for volumetric (3D) detection of the lung nodule from CT image.
This model is trained on LUNA16 dataset (https://luna16.grand-challenge.org/Home/), using the RetinaNet (Lin, Tsung-Yi, et al. "Focal loss for dense object detection." ICCV 2017. https://arxiv.org/abs/1708.02002).
Data
The dataset we are experimenting in this example is LUNA16 (https://luna16.grand-challenge.org/Home/), which is based on
LIDC-IDRI database
[3,4,5].
LUNA16 is a public dataset of CT lung nodule detection. Using raw CT scans, the goal is to identify locations of possible nodules, and to assign a probability for being a nodule to each location.
Disclaimer: We are not the host of the data. Please make sure to read the requirements and usage policies of the data and give credit to the authors of the dataset! We acknowledge the National Cancer Institute and the Foundation for the National Institutes of Health, and their critical role in the creation of the free publicly available LIDC/IDRI Database used in this study.
10-fold data splitting
We follow the official 10-fold data splitting from LUNA16 challenge and generate data split json files using the script from
nnDetection
.
Please download the resulted json files from https://github.com/Project-MONAI/MONAI-extra-test-data/releases/download/0.8.1/LUNA16_datasplit-20220615T233840Z-001.zip.
In these files, the values of "box" are the ground truth boxes in world coordinate.
Data resampling
The raw CT images in LUNA16 have various of voxel sizes. The first step is to resample them to the same voxel size.
In this model, we resampled them into 0.703125 x 0.703125 x 1.25 mm.
Please following the instruction in Section 3.1 of https://github.com/Project-MONAI/tutorials/tree/main/detection to do the resampling.
Data download
The mhd/raw original data can be downloaded from
LUNA16
. The DICOM original data can be downloaded from
LIDC-IDRI database
[3,4,5]. You will need to resample the original data to start training.
Alternatively, we provide
resampled nifti images
and a copy of
original mhd/raw images
from
LUNA16
for users to download.
Training configuration
The training was performed with the following:
-
GPU: at least 16GB GPU memory, requires 32G when exporting TRT model
-
Actual Model Input: 192 x 192 x 80
-
AMP: True
-
Optimizer: Adam
-
Learning Rate: 1e-2
-
Loss: BCE loss and L1 loss
Input
1 channel
- List of 3D CT patches
Output
In Training Mode: A dictionary of classification and box regression loss.
In Evaluation Mode: A list of dictionaries of predicted box, classification label, and classification score.
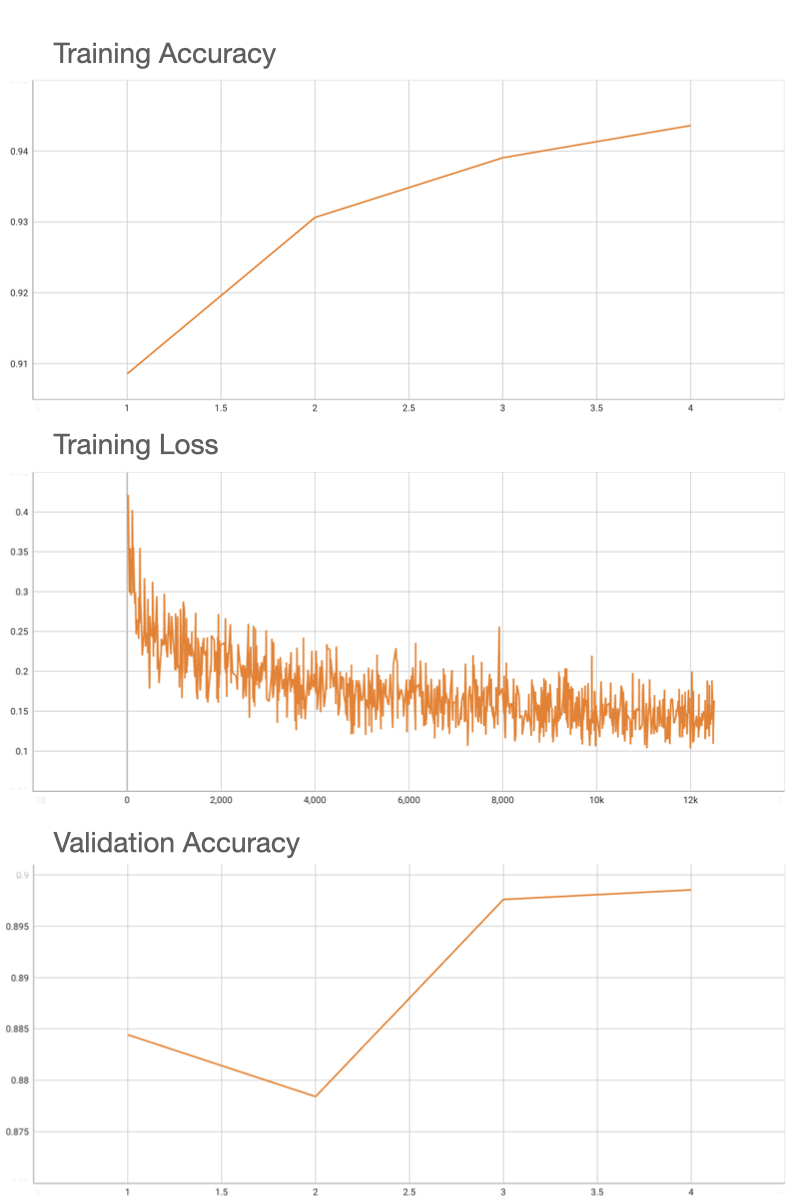
Performance
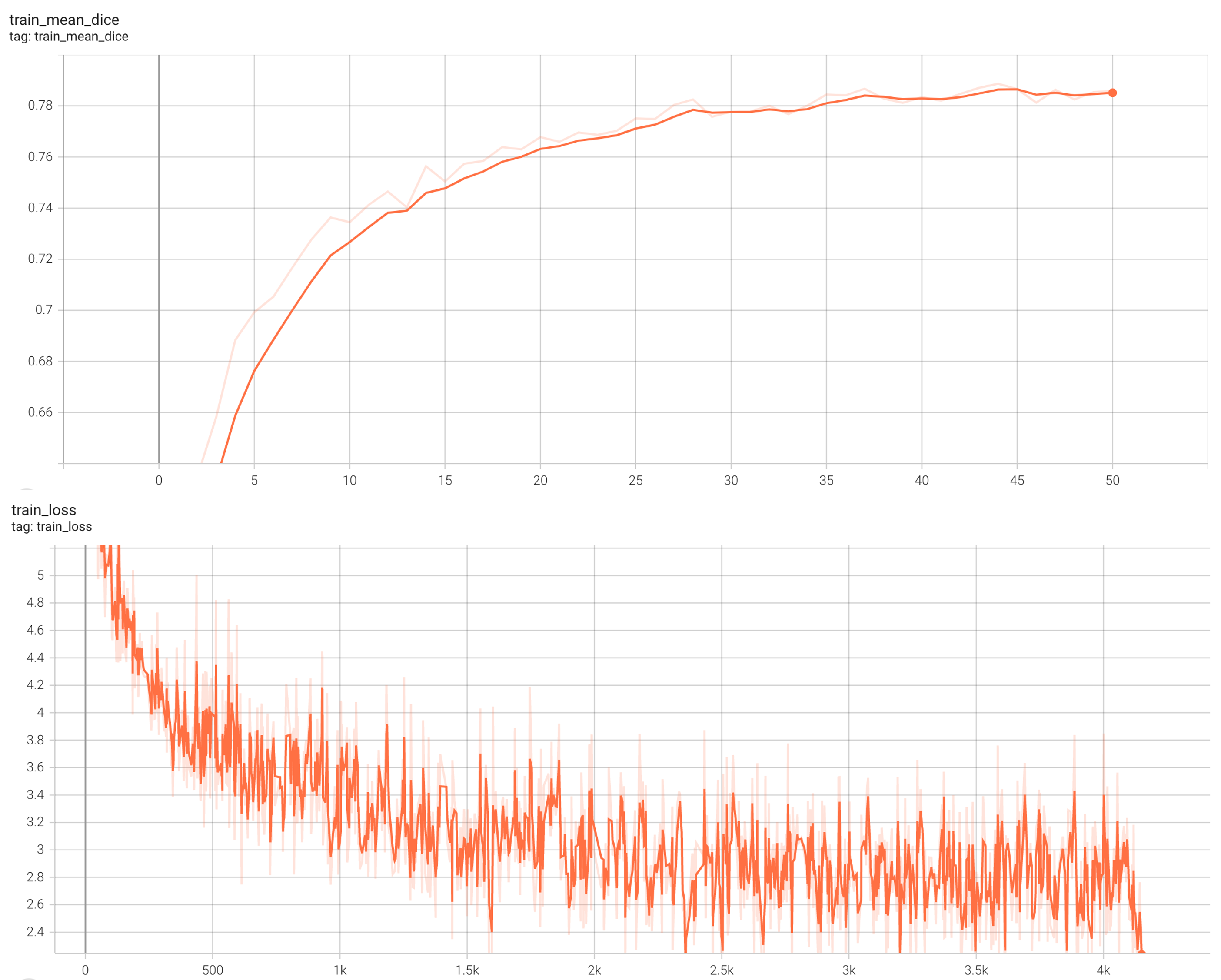
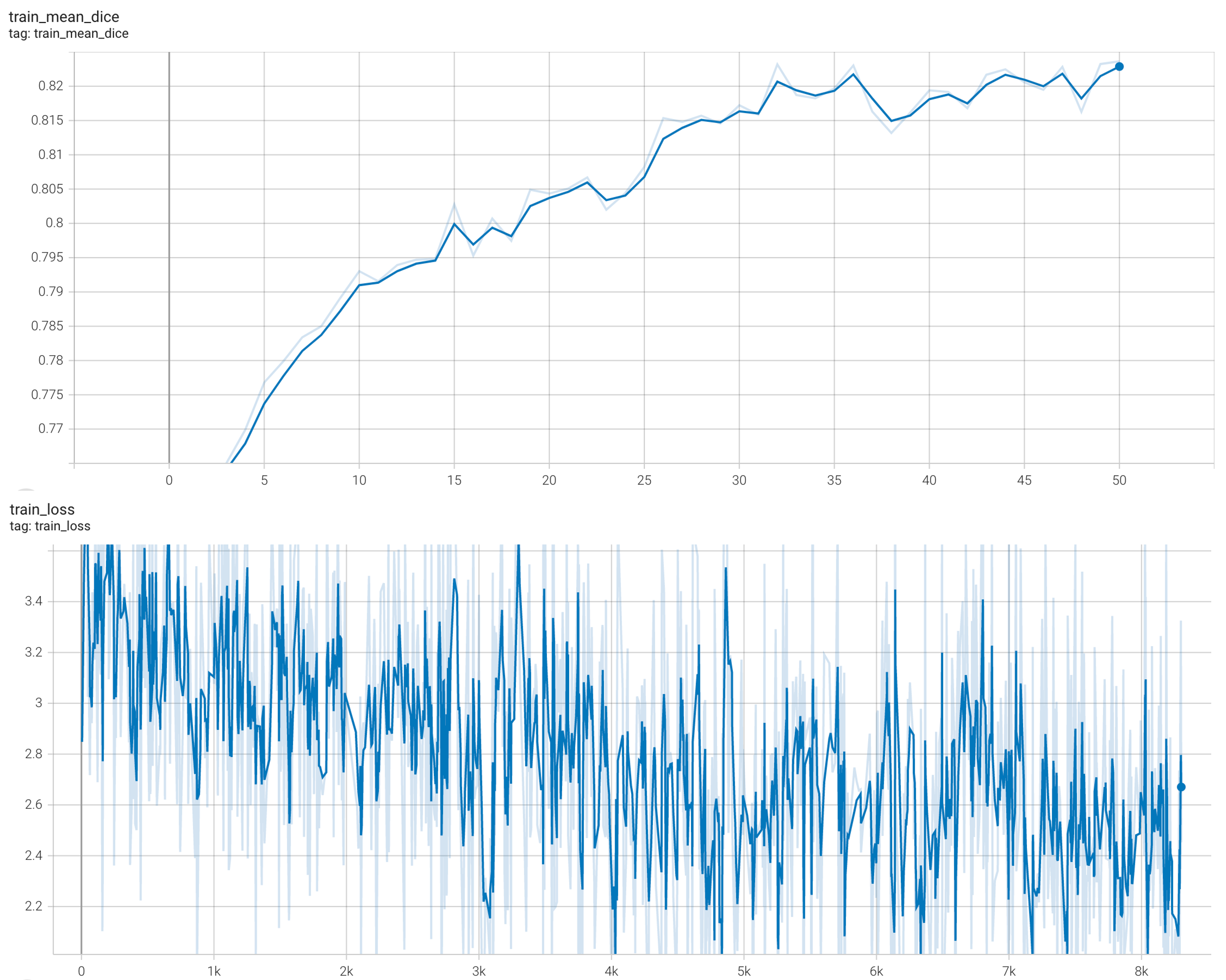
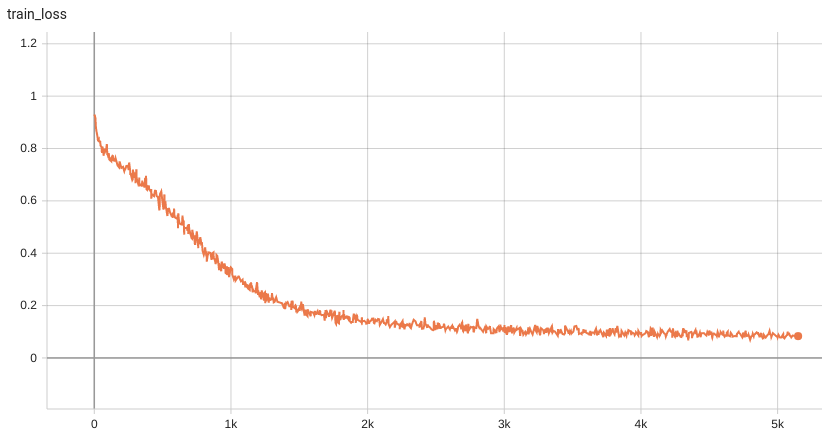
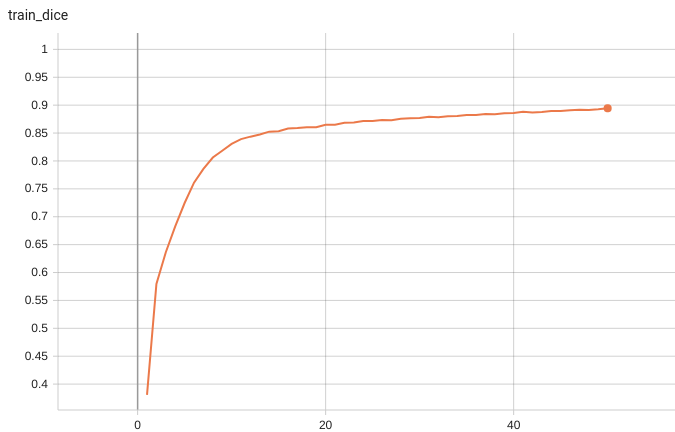
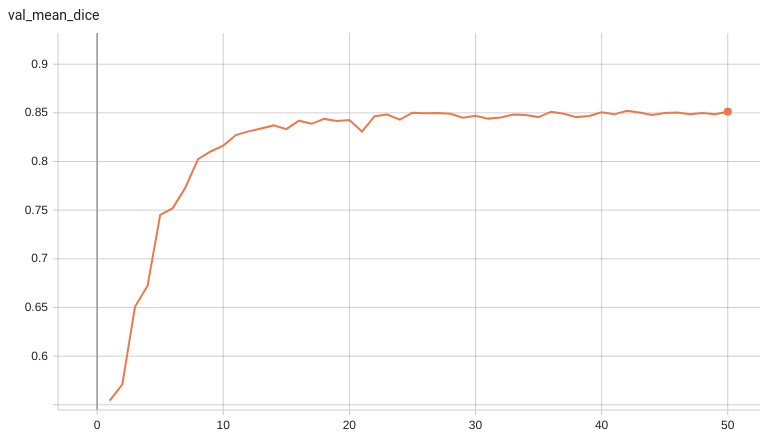
Coco metric is used for evaluating the performance of the model. The pre-trained model was trained and validated on data fold 0. This model achieves a mAP=0.852, mAR=0.998, AP(IoU=0.1)=0.858, AR(IoU=0.1)=1.0.
Please note that this bundle is non-deterministic because of the max pooling layer used in the network. Therefore, reproducing the training process may not get exactly the same performance.
Please refer to https://pytorch.org/docs/stable/notes/randomness.html#reproducibility for more details about reproducibility.
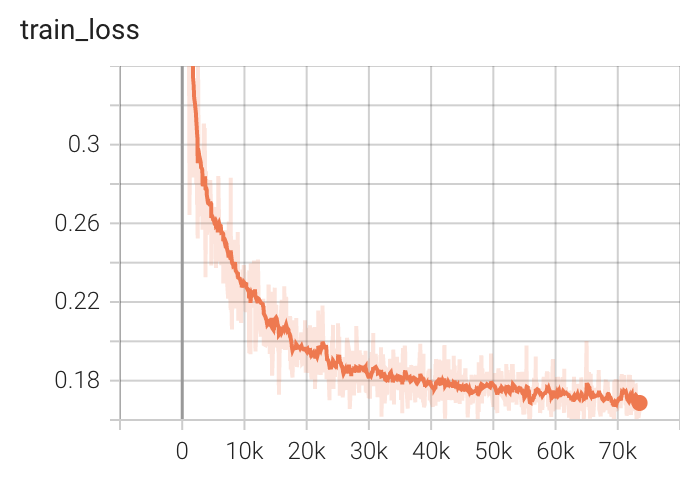
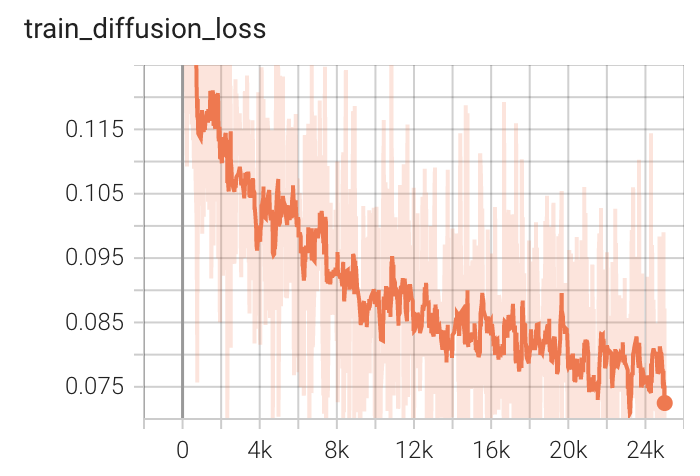
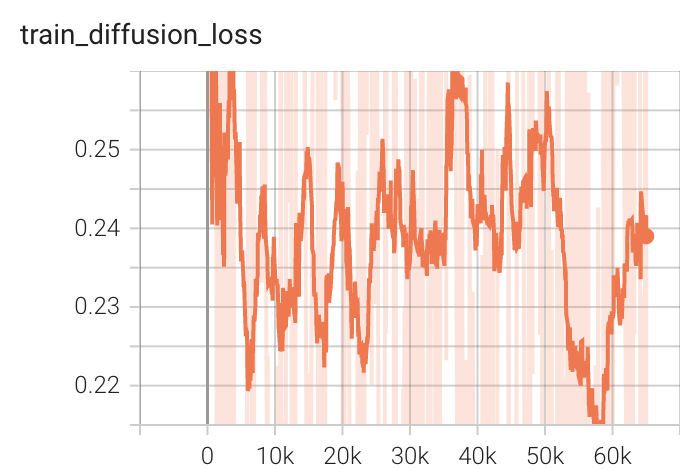
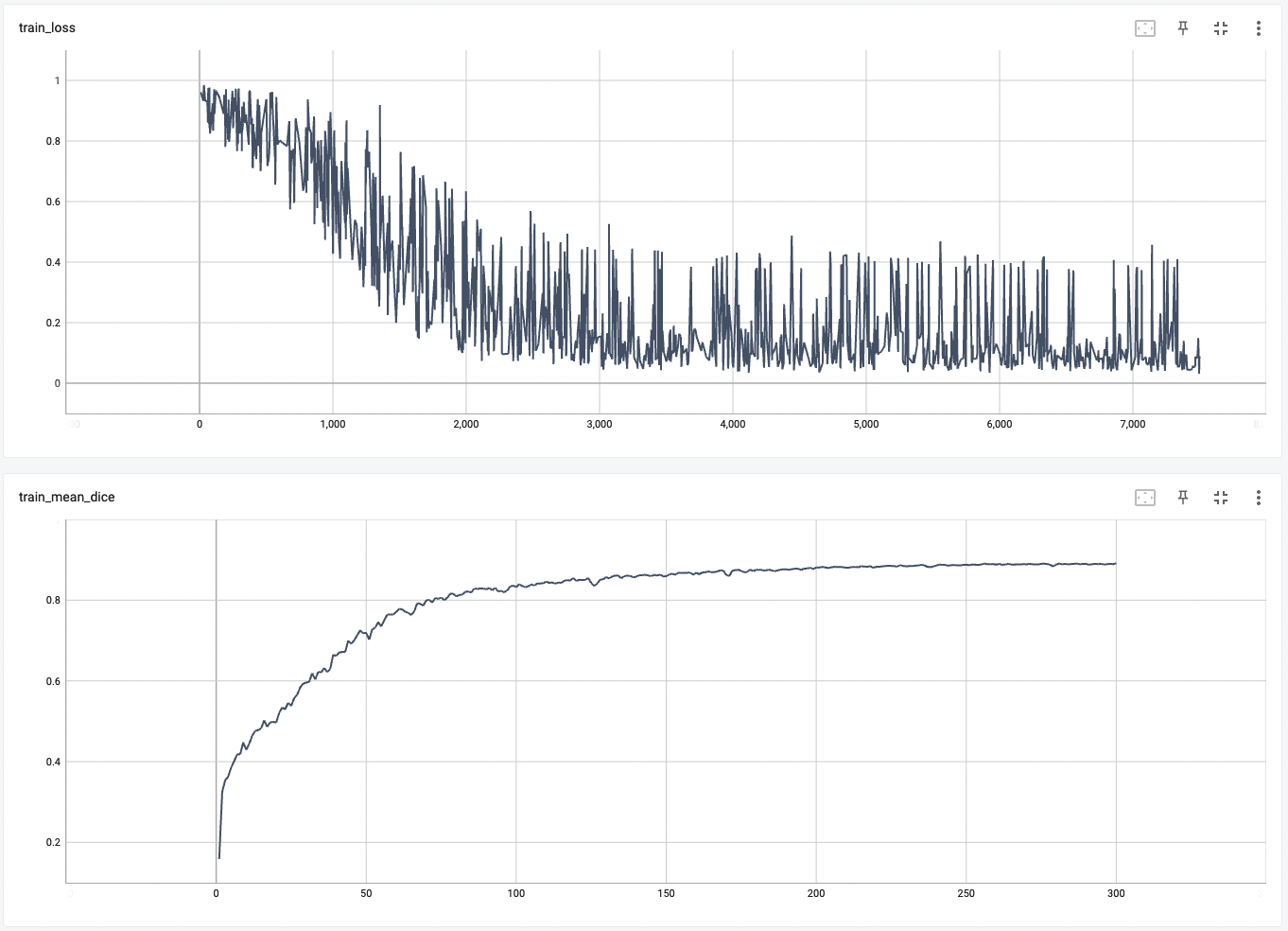
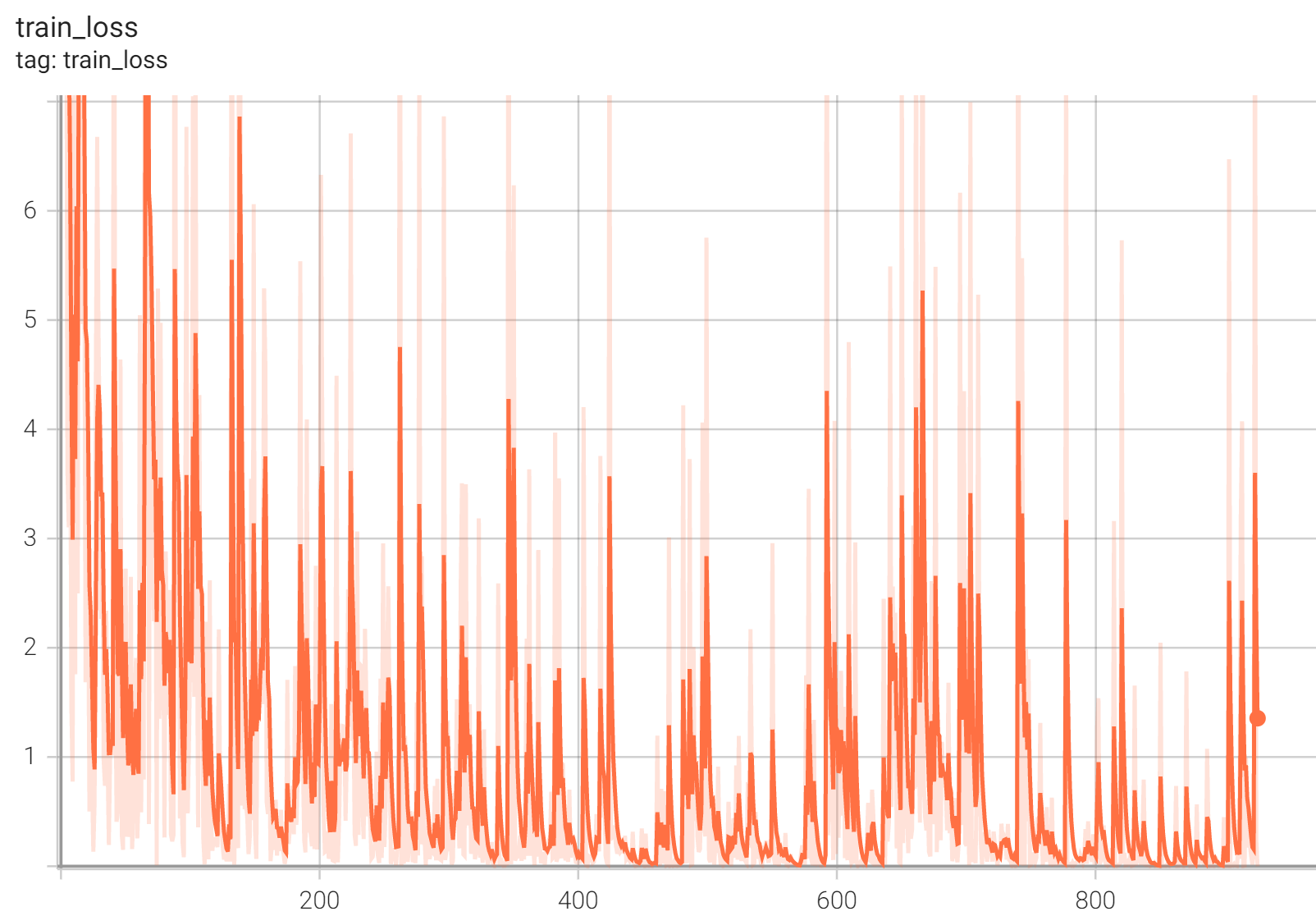
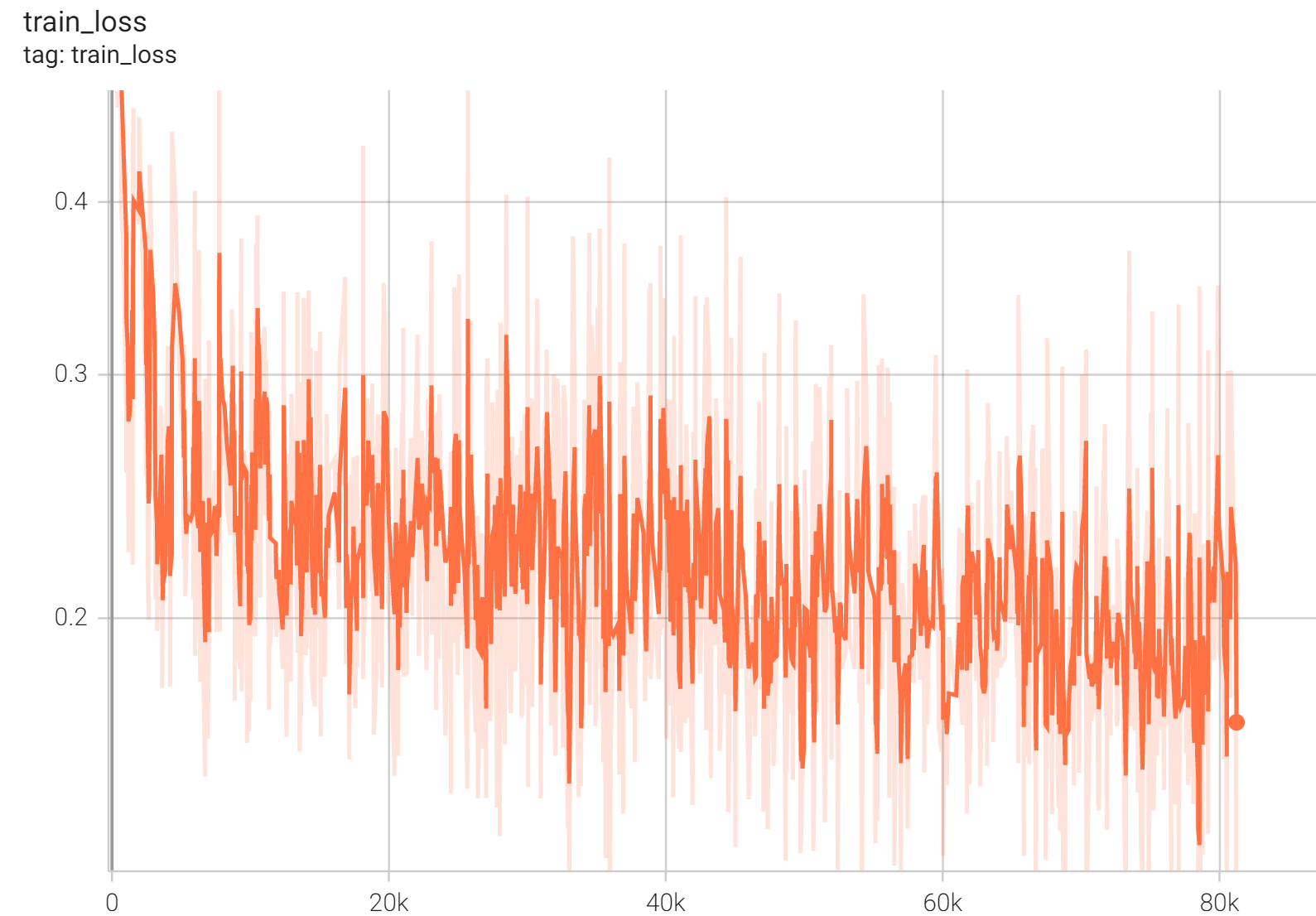
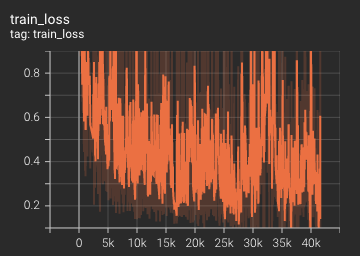
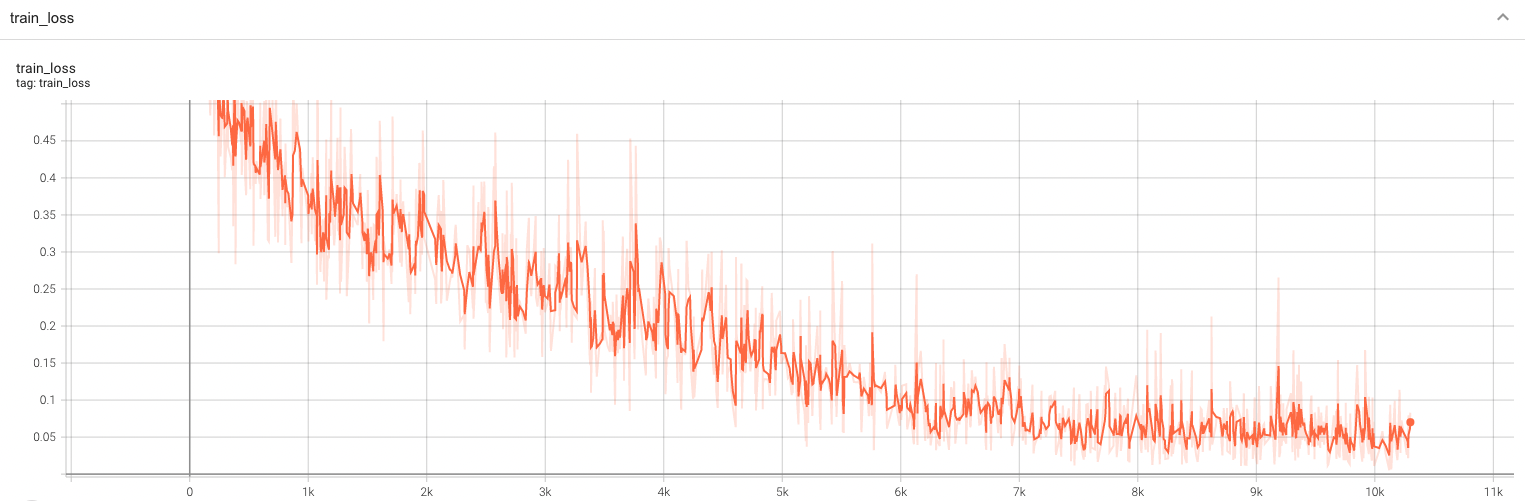
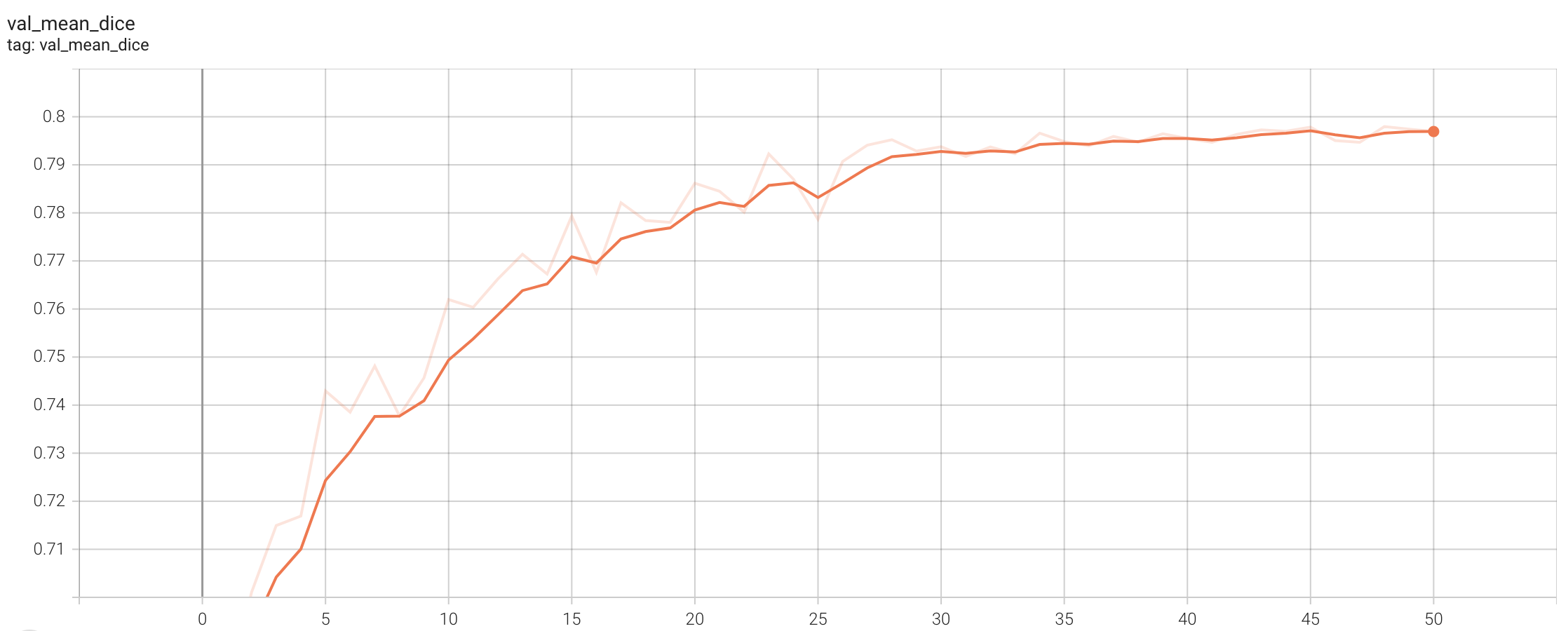
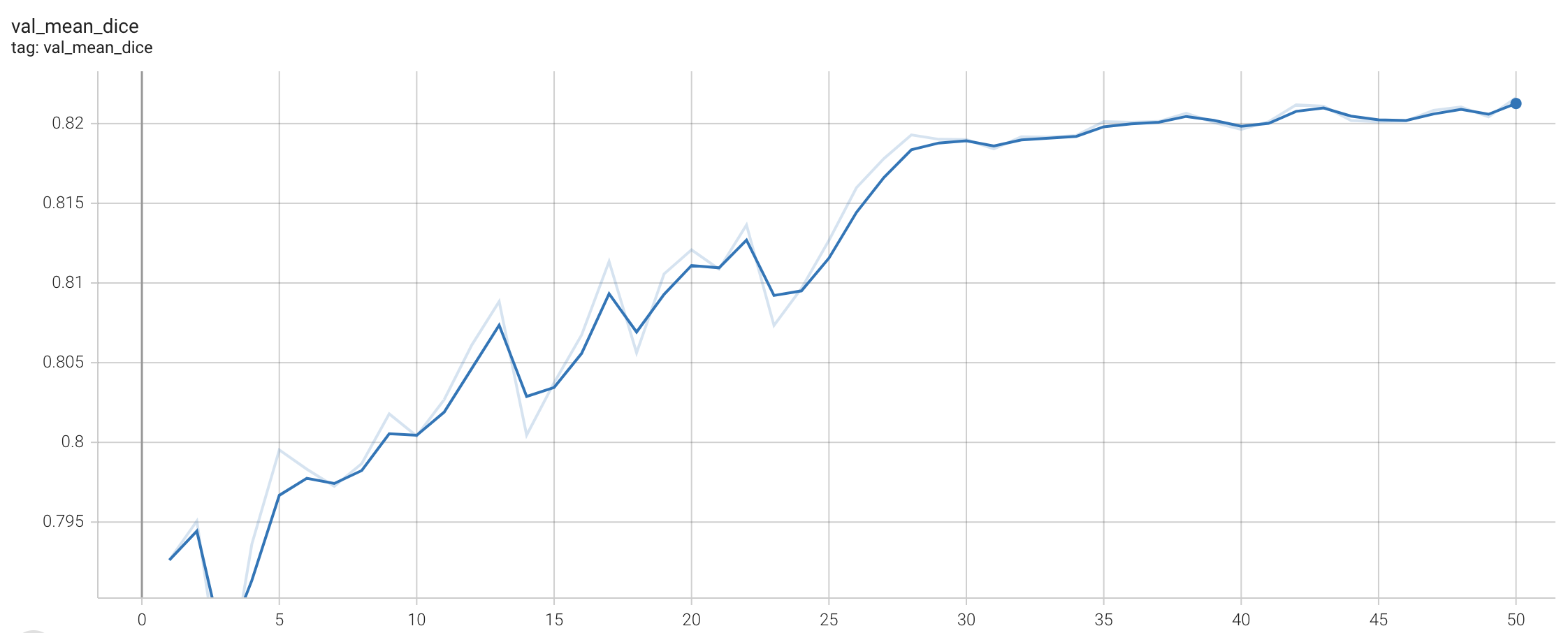
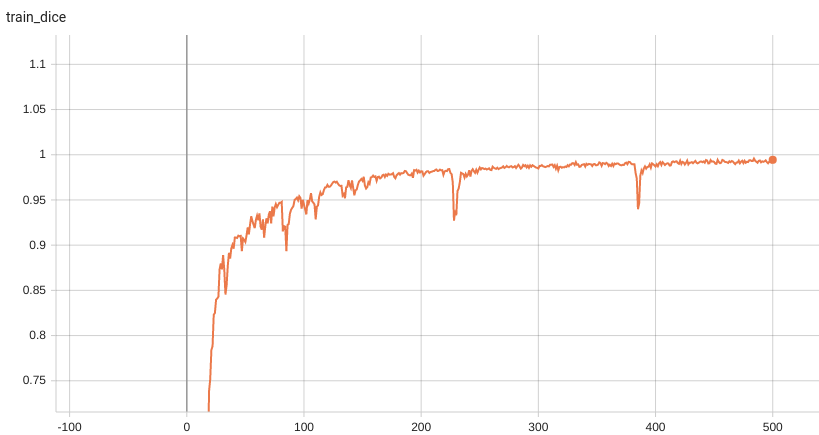
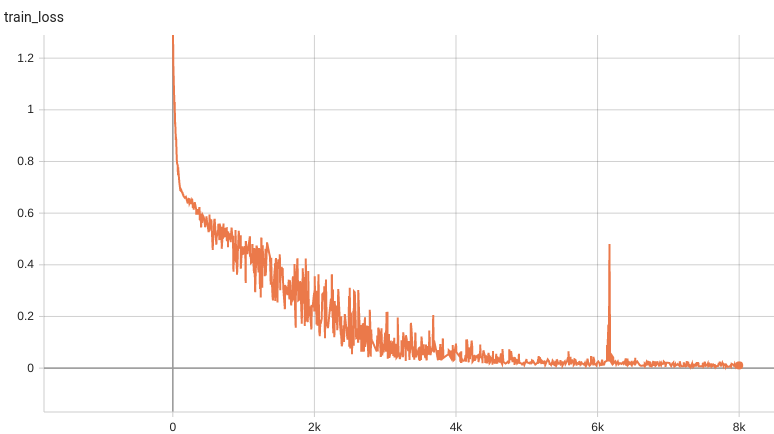
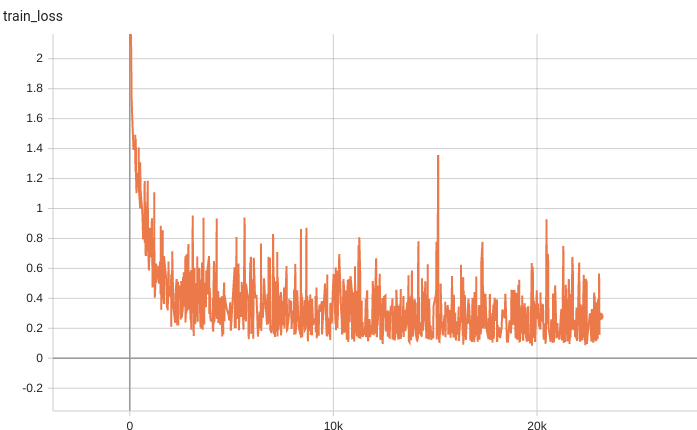
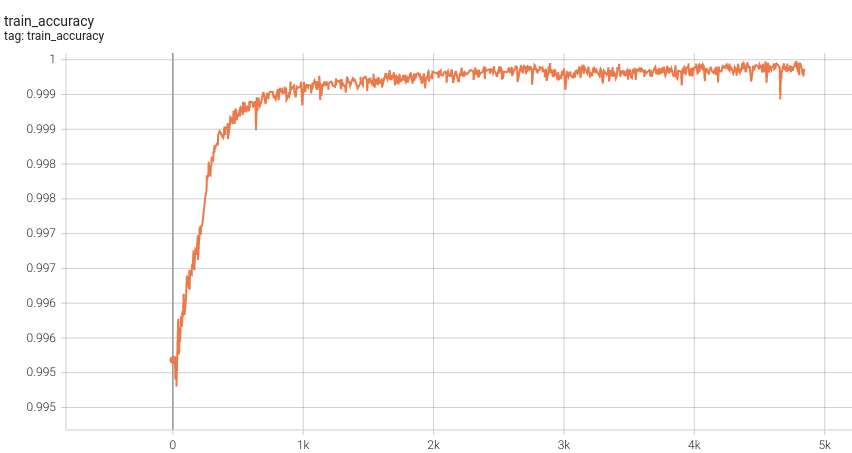
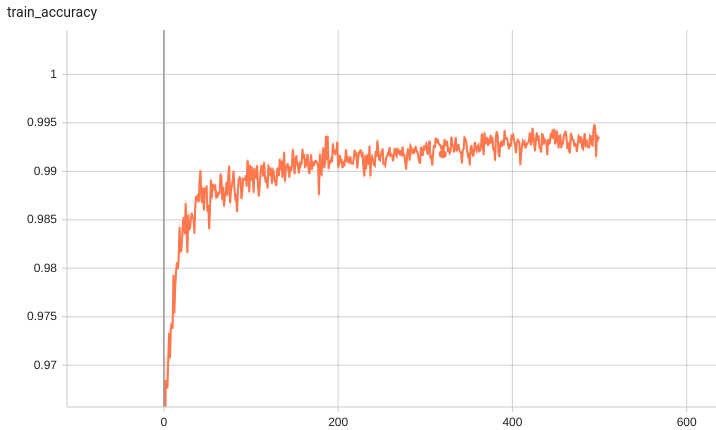
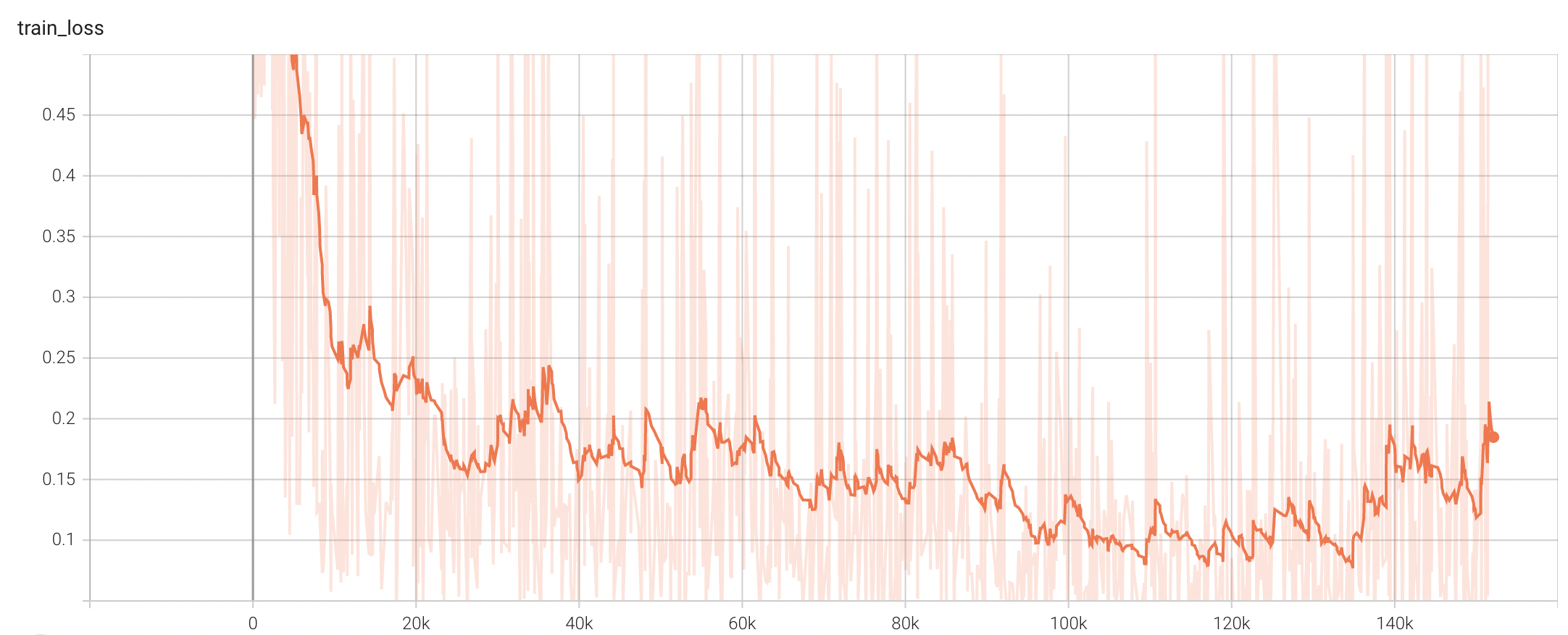
Training Loss
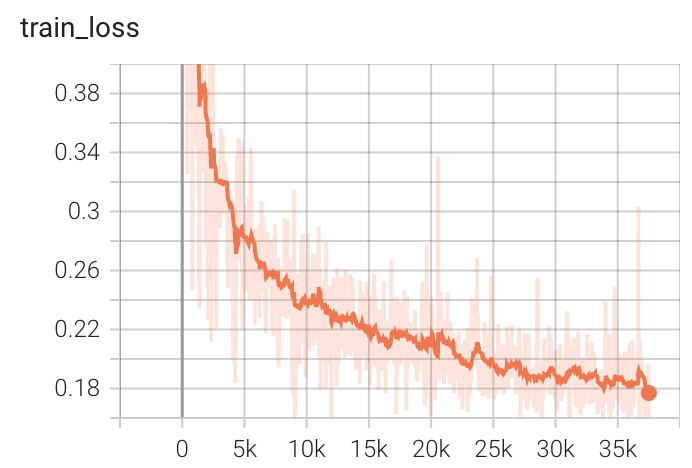
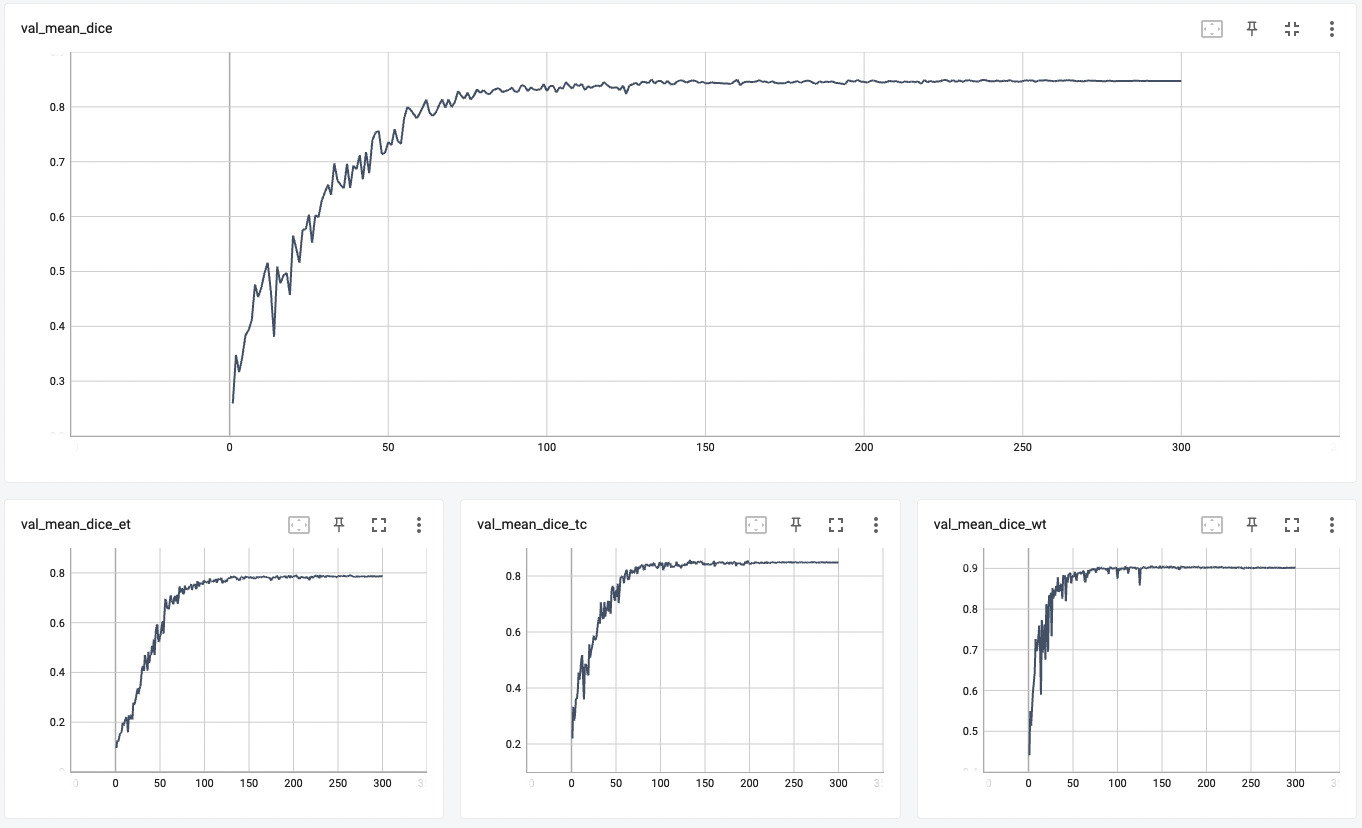
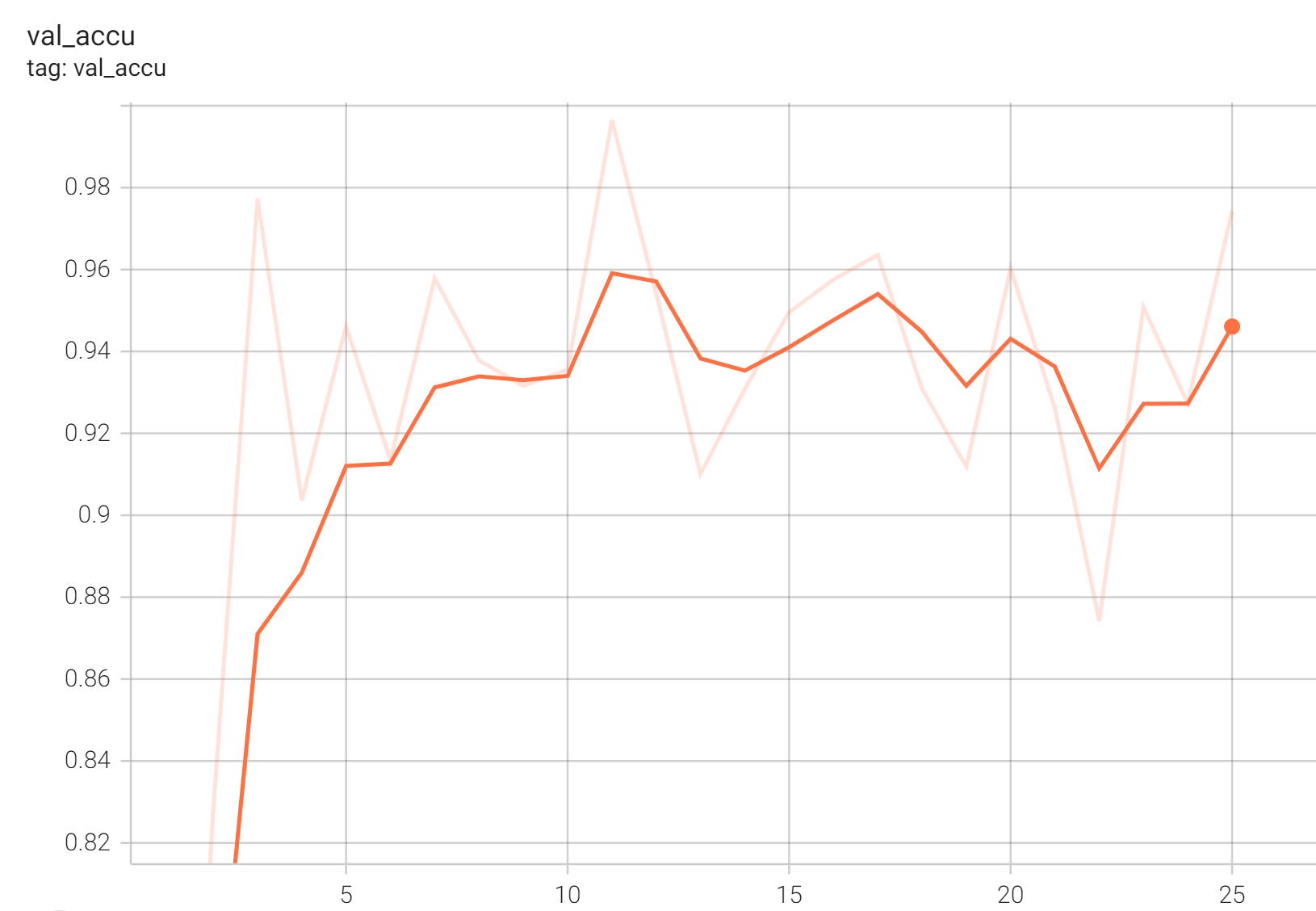
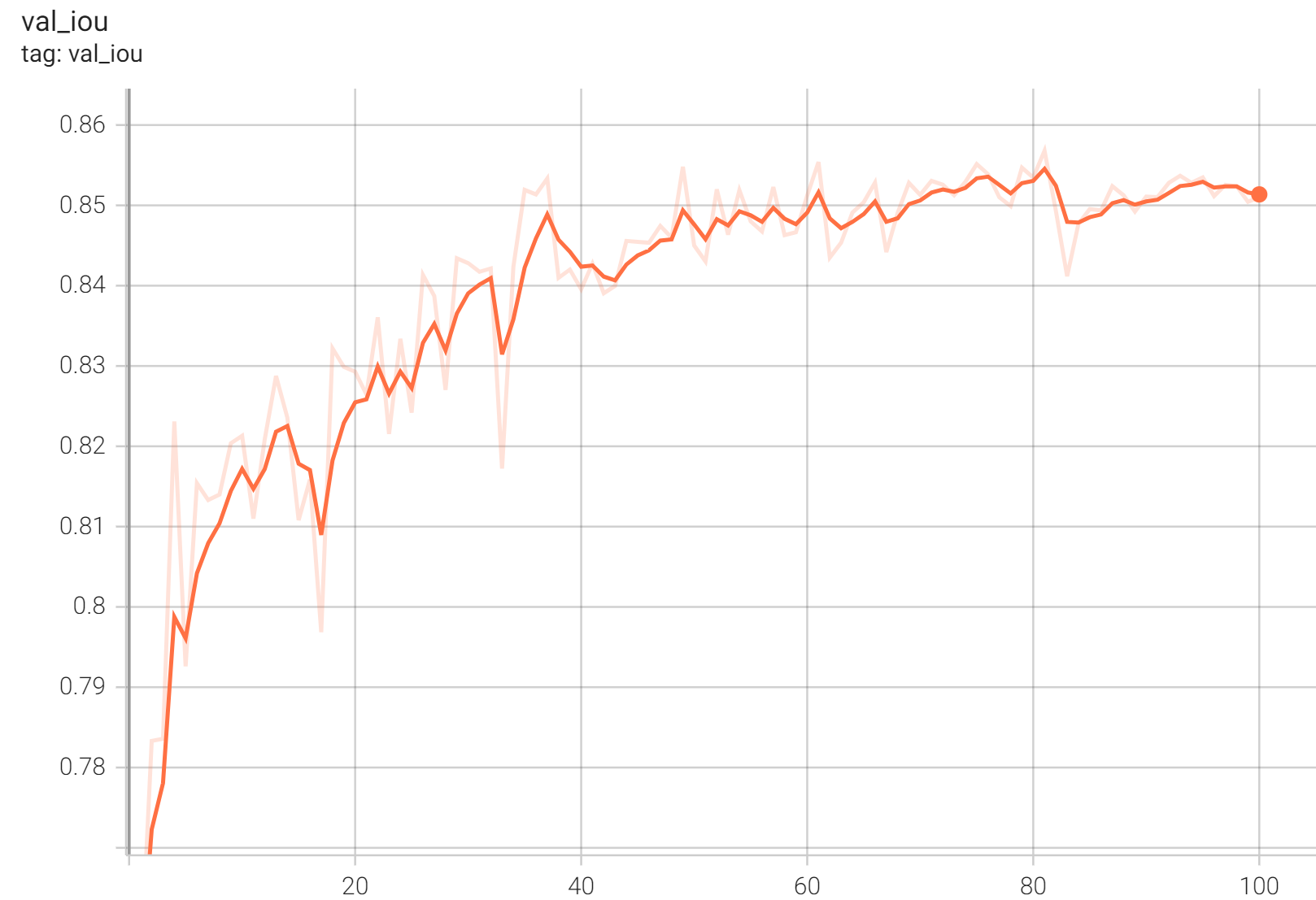
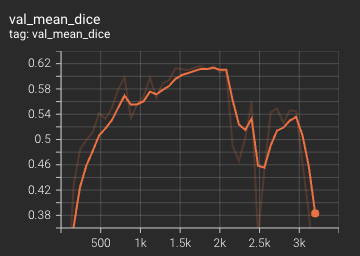
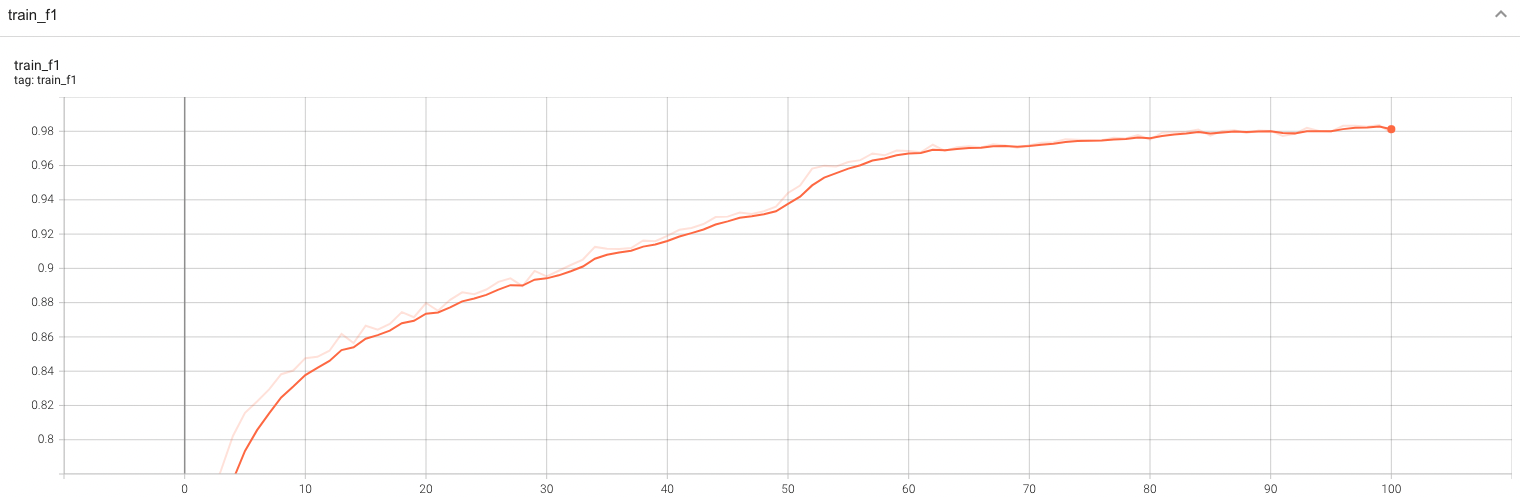
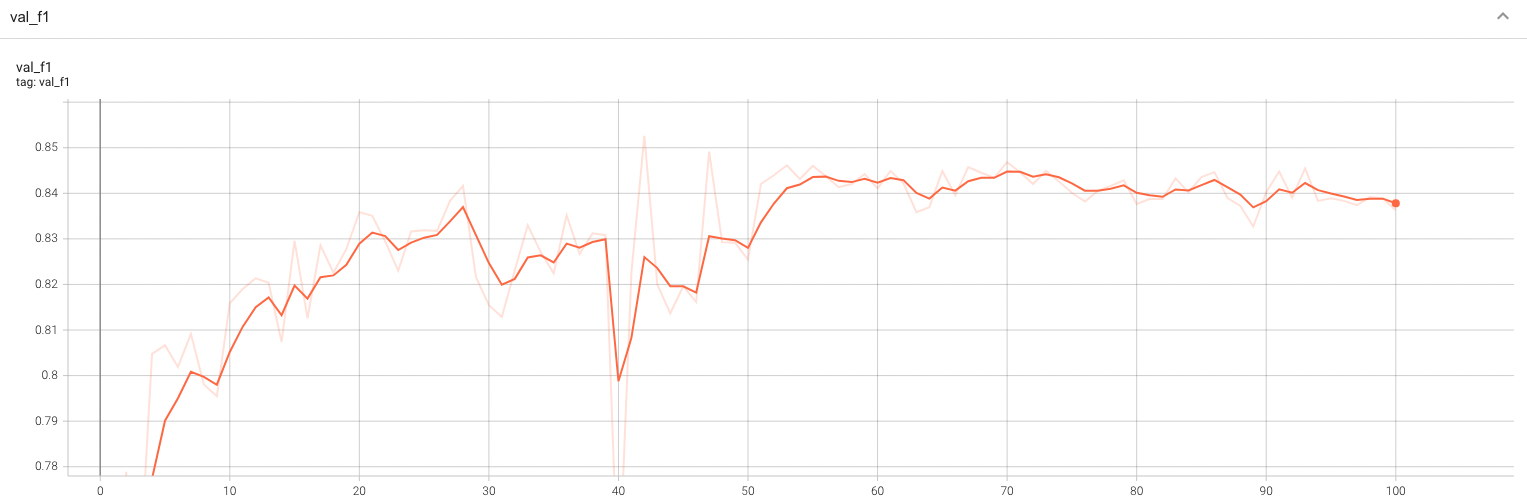
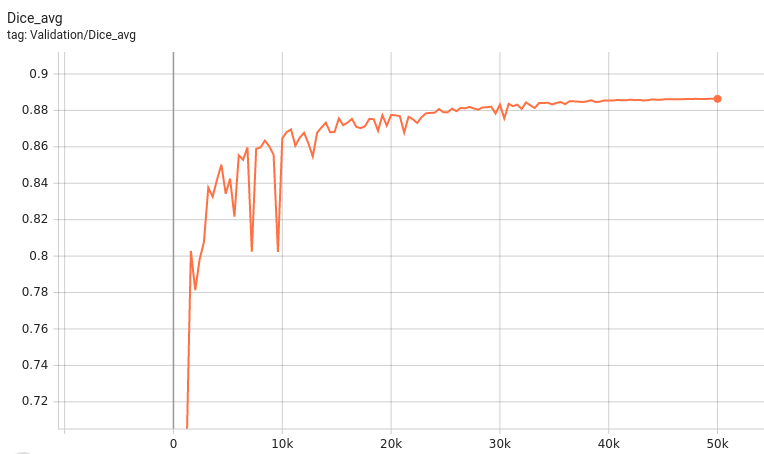
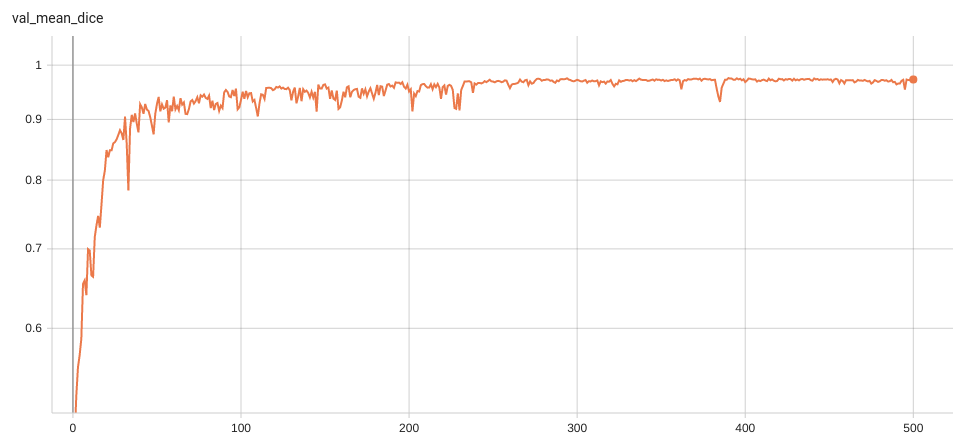
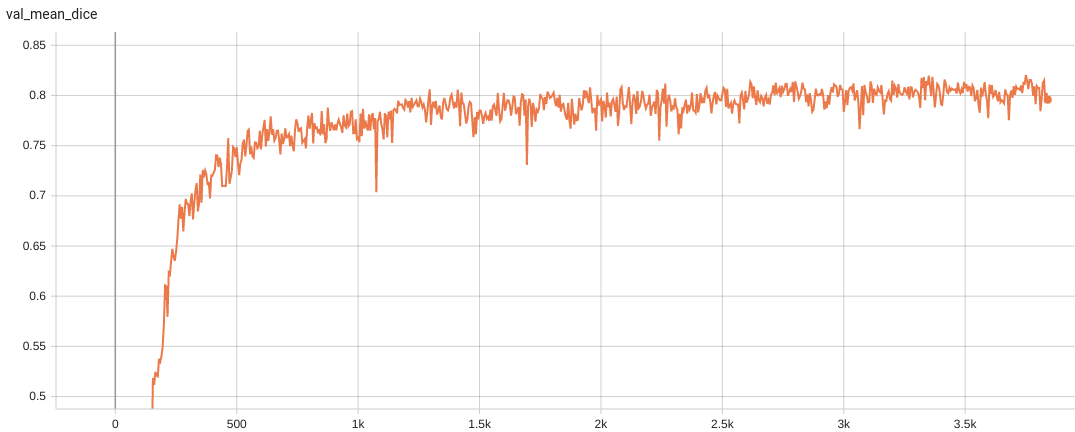
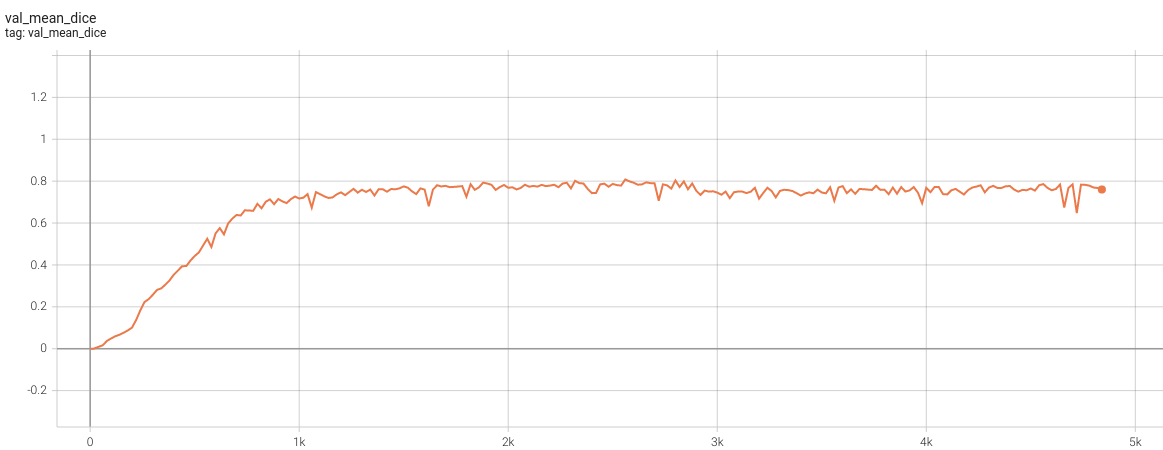
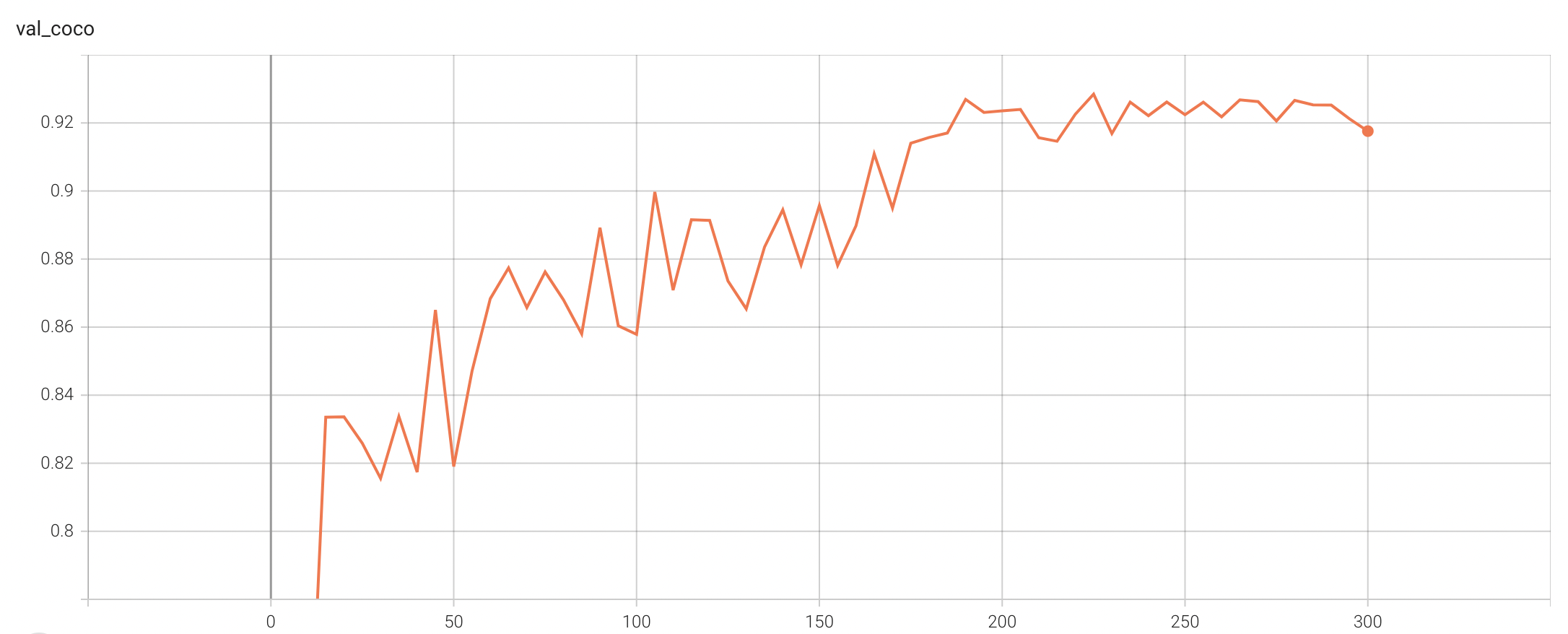
Validation Accuracy
The validation accuracy in this curve is the mean of mAP, mAR, AP(IoU=0.1), and AR(IoU=0.1) in Coco metric.
TensorRT speedup
The
lung_nodule_ct_detection
bundle supports acceleration with TensorRT through the ONNX-TensorRT method. The table below displays the speedup ratios observed on an A100 80G GPU. Please note that when using the TensorRT model for inference, the
force_sliding_window
parameter in the
inference.json
file must be set to
true
. This ensures that the bundle uses the
SlidingWindowInferer
during inference and maintains the input spatial size of the network. Otherwise, if given an input with spatial size less than the
infer_patch_size
, the input spatial size of the network would be changed.
|
method
|
torch_fp32(ms)
|
torch_amp(ms)
|
trt_fp32(ms)
|
trt_fp16(ms)
|
speedup amp
|
speedup fp32
|
speedup fp16
|
amp vs fp16
|
|
model computation
|
7449.84
|
996.08
|
976.67
|
626.90
|
7.63
|
7.63
|
11.88
|
1.56
|
|
end2end
|
36458.26
|
7259.35
|
6420.60
|
4698.34
|
5.02
|
5.68
|
7.76
|
1.55
|
Where:
-
model computation
means the speedup ratio of model's inference with a random input without preprocessing and postprocessing
-
end2end
means run the bundle end-to-end with the TensorRT based model.
-
torch_fp32
and
torch_amp
are for the PyTorch models with or without
amp
mode.
-
trt_fp32
and
trt_fp16
are for the TensorRT based models converted in corresponding precision.
-
speedup amp
,
speedup fp32
and
speedup fp16
are the speedup ratios of corresponding models versus the PyTorch float32 model
-
amp vs fp16
is the speedup ratio between the PyTorch amp model and the TensorRT float16 based model.
Currently, the only available method to accelerate this model is through ONNX-TensorRT. However, the Torch-TensorRT method is under development and will be available in the near future.
This result is benchmarked under:
- TensorRT: 8.5.3+cuda11.8
- Torch-TensorRT Version: 1.4.0
- CPU Architecture: x86-64
- OS: ubuntu 20.04
- Python version:3.8.10
- CUDA version: 12.0
- GPU models and configuration: A100 80G
MONAI Bundle Commands
In addition to the Pythonic APIs, a few command line interfaces (CLI) are provided to interact with the bundle. The CLI supports flexible use cases, such as overriding configs at runtime and predefining arguments in a file.
For more details usage instructions, visit the
MONAI Bundle Configuration Page
.
Execute training:
python -m monai.bundle run --config_file configs/train.json
Please note that if the default dataset path is not modified with the actual path in the bundle config files, you can also override it by using
--dataset_dir
:
python -m monai.bundle run --config_file configs/train.json --dataset_dir <actual dataset path>
Override the
train
config to execute evaluation with the trained model:
python -m monai.bundle run --config_file "['configs/train.json','configs/evaluate.json']"
Execute inference on resampled LUNA16 images by setting
"whether_raw_luna16": false
in
inference.json
:
python -m monai.bundle run --config_file configs/inference.json
With the same command, we can execute inference on original LUNA16 images by setting
"whether_raw_luna16": true
in
inference.json
. Remember to also set
"data_list_file_path": "$@bundle_root + '/LUNA16_datasplit/mhd_original/dataset_fold0.json'"
and change
"dataset_dir"
.
Note that in inference.json, the transform "LoadImaged" in "preprocessing" and "AffineBoxToWorldCoordinated" in "postprocessing" has
"affine_lps_to_ras": true
.
This depends on the input images. LUNA16 needs
"affine_lps_to_ras": true
.
It is possible that your inference dataset should set
"affine_lps_to_ras": false
.
Export checkpoint to TensorRT based models with fp32 or fp16 precision
python -m monai.bundle trt_export --net_id network_def --filepath models/model_trt.ts --ckpt_file models/model.pt --meta_file configs/metadata.json --config_file configs/inference.json --precision <fp32/fp16> --input_shape "[1, 1, 512, 512, 192]" --use_onnx "True" --use_trace "True" --onnx_output_names "['output_0', 'output_1', 'output_2', 'output_3', 'output_4', 'output_5']" --network_def#use_list_output "True"
Execute inference with the TensorRT model
python -m monai.bundle run --config_file "['configs/inference.json', 'configs/inference_trt.json']"
References
[1] Lin, Tsung-Yi, et al. "Focal loss for dense object detection." ICCV 2017. https://arxiv.org/abs/1708.02002)
[2] Baumgartner and Jaeger et al. "nnDetection: A self-configuring method for medical object detection." MICCAI 2021. https://arxiv.org/pdf/2106.00817.pdf
[3] Armato III, S. G., McLennan, G., Bidaut, L., McNitt-Gray, M. F., Meyer, C. R., Reeves, A. P., Zhao, B., Aberle, D. R., Henschke, C. I., Hoffman, E. A., Kazerooni, E. A., MacMahon, H., Van Beek, E. J. R., Yankelevitz, D., Biancardi, A. M., Bland, P. H., Brown, M. S., Engelmann, R. M., Laderach, G. E., Max, D., Pais, R. C. , Qing, D. P. Y. , Roberts, R. Y., Smith, A. R., Starkey, A., Batra, P., Caligiuri, P., Farooqi, A., Gladish, G. W., Jude, C. M., Munden, R. F., Petkovska, I., Quint, L. E., Schwartz, L. H., Sundaram, B., Dodd, L. E., Fenimore, C., Gur, D., Petrick, N., Freymann, J., Kirby, J., Hughes, B., Casteele, A. V., Gupte, S., Sallam, M., Heath, M. D., Kuhn, M. H., Dharaiya, E., Burns, R., Fryd, D. S., Salganicoff, M., Anand, V., Shreter, U., Vastagh, S., Croft, B. Y., Clarke, L. P. (2015). Data From LIDC-IDRI [Data set]. The Cancer Imaging Archive. https://doi.org/10.7937/K9/TCIA.2015.LO9QL9SX
[4] Armato SG 3rd, McLennan G, Bidaut L, McNitt-Gray MF, Meyer CR, Reeves AP, Zhao B, Aberle DR, Henschke CI, Hoffman EA, Kazerooni EA, MacMahon H, Van Beeke EJ, Yankelevitz D, Biancardi AM, Bland PH, Brown MS, Engelmann RM, Laderach GE, Max D, Pais RC, Qing DP, Roberts RY, Smith AR, Starkey A, Batrah P, Caligiuri P, Farooqi A, Gladish GW, Jude CM, Munden RF, Petkovska I, Quint LE, Schwartz LH, Sundaram B, Dodd LE, Fenimore C, Gur D, Petrick N, Freymann J, Kirby J, Hughes B, Casteele AV, Gupte S, Sallamm M, Heath MD, Kuhn MH, Dharaiya E, Burns R, Fryd DS, Salganicoff M, Anand V, Shreter U, Vastagh S, Croft BY. The Lung Image Database Consortium (LIDC) and Image Database Resource Initiative (IDRI): A completed reference database of lung nodules on CT scans. Medical Physics, 38: 915--931, 2011. DOI: https://doi.org/10.1118/1.3528204
[5] Clark, K., Vendt, B., Smith, K., Freymann, J., Kirby, J., Koppel, P., Moore, S., Phillips, S., Maffitt, D., Pringle, M., Tarbox, L., & Prior, F. (2013). The Cancer Imaging Archive (TCIA): Maintaining and Operating a Public Information Repository. Journal of Digital Imaging, 26(6), 1045–1057. https://doi.org/10.1007/s10278-013-9622-7
License
Copyright (c) MONAI Consortium
Licensed under the Apache License, Version 2.0 (the "License");
you may not use this file except in compliance with the License.
You may obtain a copy of the License at
http://www.apache.org/licenses/LICENSE-2.0
Unless required by applicable law or agreed to in writing, software
distributed under the License is distributed on an "AS IS" BASIS,
WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
See the License for the specific language governing permissions and
limitations under the License.