MONAI Label
MONAI Label is an intelligent image labeling and learning tool that enables you to create training datasets and build AI annotation models to accelerate the development of AI applications in medical imaging.
Annotation Tools for Clinical Workflows
MONAI Label combines AI assistance with clinical expertise to deliver fast, accurate, and consistent annotations. Our tools adapt to your workflow while continuously learning from your expertise.
AI-Assisted Annotation
Interactive AI models for faster annotation
Active learning for model improvement
Real-time AI assistance
Clinical Integration
3D Slicer, MITK, OHIF viewer integration
DICOM and PACS integration
Multi-user collaboration support
Extensible Platform
Customizable annotation strategies
Plugin architecture for flexibility
Easy integration with existing tools
Domain-Specific Solutions
MONAI Label delivers specialized solutions optimized for each medical imaging domain. Our tools adapt to your specific needs, whether you're working with 3D volumes, microscopy slides, or video sequences.
Radiology
Perfect for organ segmentation, tumor detection, and anatomical measurements in CT and MRI scans. Seamlessly integrated with OHIF, MITK, and 3D Slicer, our tools accelerate volumetric analysis while maintaining high accuracy.
Pathology
Specialized in cell detection and tissue classification for whole slide imaging. Integrated with QuPath, DSA, and CellProfiler for rapid analysis of microscopy data.
Endoscopy
Optimized for polyp detection and tool tracking in video sequences. Integrated with CVAT for efficient frame-by-frame annotation with automated tracking.
Getting Started with MONAI Label
Get started with MONAI Label in minutes. Our step-by-step guide will help you set up your environment and launch your first AI-assisted annotation server. After setup, you'll be ready to connect to your preferred viewer and start annotating with AI assistance.
Install MONAI Label
Install via pip package manager
Download Sample App
Get a pre-configured radiology application
Download Sample Dataset
Get example medical imaging data
Launch Server
Start MONAI Label server with your configuration
Intelligent Active Learning
MONAI Label's intelligent active learning system dramatically reduces annotation time while maintaining high accuracy. Our advanced algorithms identify the most informative samples and continuously improve model performance.
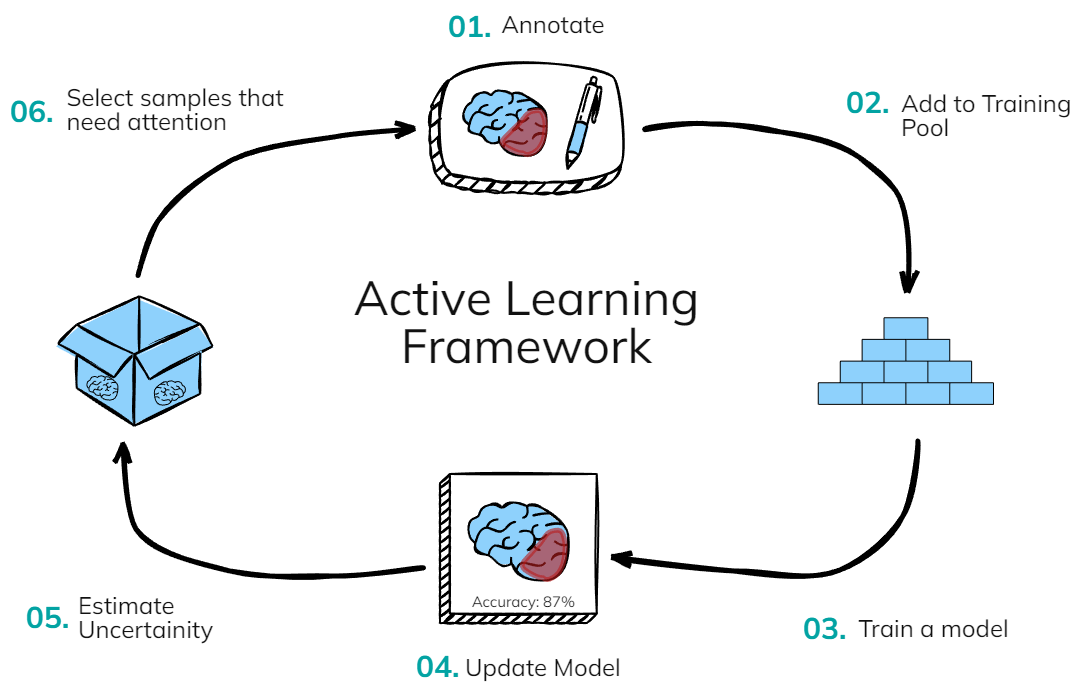
Faster annotation time
Faster model convergence
Accuracy with fewer labels
Smart Sample Selection
- Uncertainty sampling identifies challenging cases
- Diversity metrics ensure varied training data
- Model ensemble disagreement detection
Continuous Model Improvement
- Real-time model updates during annotation
- Transfer learning from pre-trained models
- Support for UNet, UNETR, and SwinUNETR
Quality Assurance
- Real-time validation metrics
- Uncertainty estimation for predictions
- Automated annotation quality checks
Resources & Training
Get started with MONAI Label through our comprehensive learning resources and training materials.
How to Cite MONAI Label
If you use MONAI Label in your research, please cite our paper:
@article{monailabel2024,
title={MONAI Label: A framework for AI-assisted Interactive Labeling of 3D Medical Images},
author={Diaz-Pinto, Andres and Alle, Sachidanand and Nath, Vishwesh and Tang, Yucheng and Ihsani, Alvin and Asad, Muhammad and P{\'e}rez-Garc{\'i}a, Fernando and Mehta, Pritesh and Li, Wenqi and Flores, Mona and Roth, Holger R. and Vercauteren, Tom and Xu, Daguang and Dogra, Prerna and Ourselin, Sebastien and Feng, Andrew and Cardoso, M. Jorge},
journal={Medical Image Analysis},
year={2024},
doi={10.1016/j.media.2024.103207}
}
Get Involved in the Community
Join our growing community of researchers, developers, and healthcare professionals. Get help, share your work, and contribute to advancing medical image annotation. Consider joining our Human-AI Interaction Working Group to help standardize and improve AI-assisted annotation workflows.
Documentation
Access comprehensive documentation covering everything from basic concepts to advanced annotation strategies.
Human-AI Interaction Working Group
Join our working group focused on standardizing and supporting Human-AI interaction cycles through well-defined interfaces for AI-assisted annotation.
GitHub Repository
Explore the source code, contribute to development, and access sample applications for radiology, pathology, and endoscopy to learn from real-world annotation scenarios.
Join the Discussion
Connect with the MONAI community on Slack. Get help, share ideas, and collaborate with other developers.