

From Institutional Development to Scalable Deployment
Center for Augmented Intelligence in Imaging (CAII), Mayo Clinic Florida
Leveraging MONAI Deploy and Siemens Digital Marketplace for Global Research Access
Introduction
At the Mayo Clinic's Center for Augmented Intelligence in Imaging (CAII), we develop AI applications to address unmet needs in imaging research. While internal development and deployment offer opportunities to validate and refine these tools, bringing them to external research environments remains a critical barrier.
This case study outlines how an AI application developed at Mayo Clinic for research purposes—using MONAI Core and MONAI Deploy—was made available to researchers at over 10,000 institutions via the Siemens Healthineers Digital Marketplace. By combining open-source tooling with scalable distribution infrastructure, this approach provides a blueprint for both developers and institutions seeking to build and access imaging AI research tools.
From Local Pipelines to Global Access
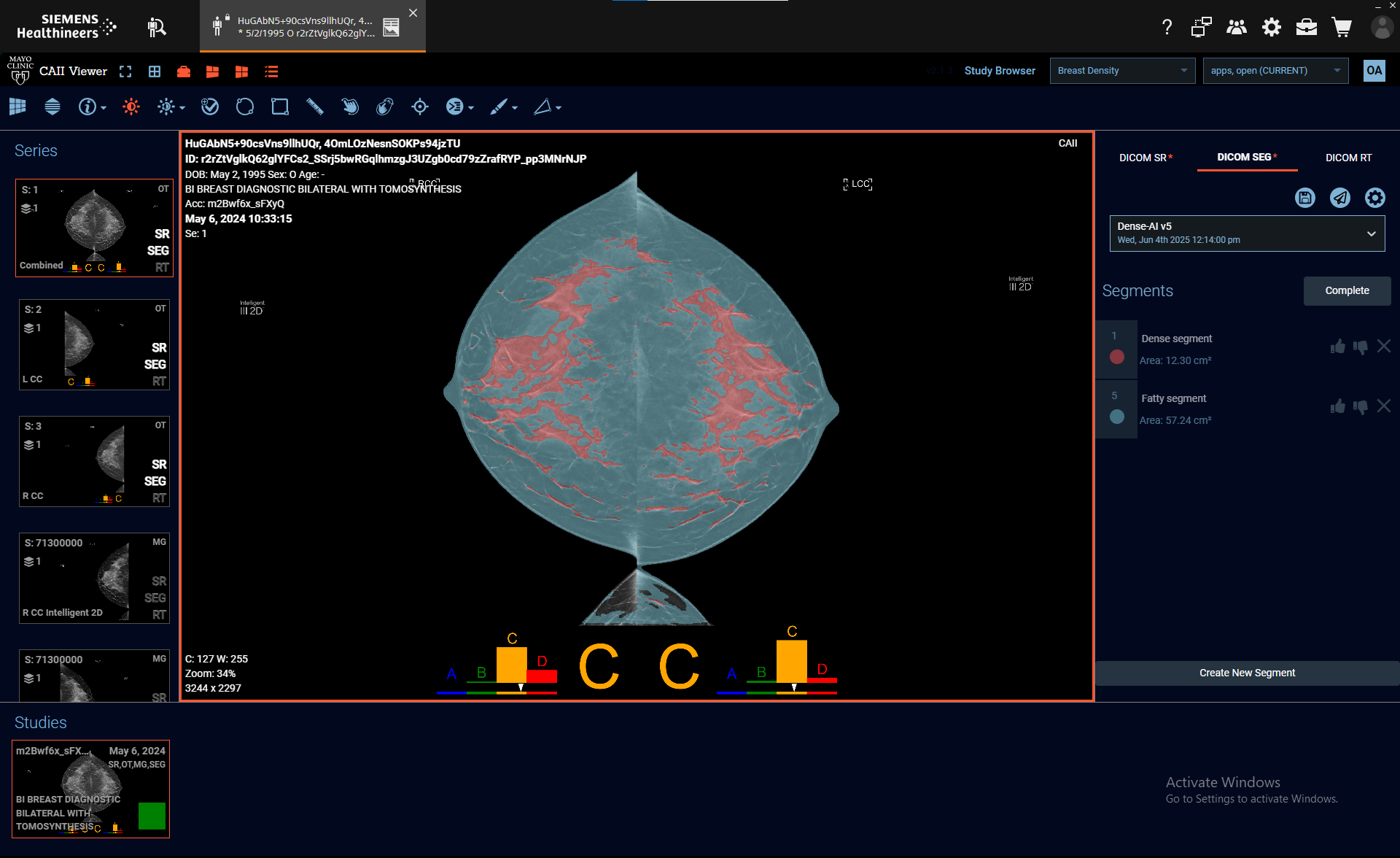
Internally, our team built and validated the AI application using MONAI for model implementation, data preprocessing, and output standardization. We relied on container-based orchestration and DICOMweb compatible CAII Viewer for integration into our clinical workflows.
However, this technical stack—while effective in-house—could not be assumed to operate at other sites. Our challenge was to preserve model reproducibility while eliminating deployment overhead for external users. We needed a delivery mechanism that allowed other institutions to run the tool locally, without requiring development experience, custom infrastructure, or vendor intervention.
The Siemens Healthineers Digital Marketplace
To address this challenge, we collaborated with Siemens Healthineers to publish the application through their Digital Marketplace—a platform designed to distribute containerized research applications for use within institutional imaging environments.
By wrapping the application in a Marketplace-compatible container and leveraging the platform's built-in rule configuration and DICOM-native integration, we enabled:
- Local execution at each site, preserving data governance
- Zero-code installation and configuration, via GUI-based tools
- DICOM-native input/output, supporting PACS integration and traceability
- Reusability across research applications, regardless of modality or model specifics
All application logic, preprocessing, visualization, and data export are bundled and versioned. The application is clearly labeled for research use only.
Developer Workflow
To assist developers aiming to bring their applications to the Digital Marketplace, Siemens Healthineers provides technical guidelines covering containerization, interface integration, and DICOM conformance. Developers are encouraged to align with MONAI Deploy App SDK structure and compatibility standards. While source code remains institutionally governed, the externalization process emphasizes modularity, traceability, and platform readiness.
Further developer resources and templates can be found at:
The developer workflow for externalization follows a clear path:
- 1. Model development and validation using MONAI Core (e.g., training, evaluation, DICOM output preparation)
- 2. Application packaging via MONAI Deploy App SDK
- 3. Containerization and alignment with Siemens Digital Marketplace standards
- 4. Integration of viewer and rule logic, using DICOMweb compatible or web-native tools
- 5. Submission and publication, including metadata, version control, and usage restrictions
Once published, the application becomes discoverable to registered institutions and can be deployed without code modifications or site-specific engineering.
User Workflow and Data Flow
For research users, the process is equally streamlined:
- 1. Discover the application in the Siemens Healthineers Digital Marketplace
- 2. Install via the syngo.via OpenApps interface or equivalent local platform
- 3. Configure rules (e.g., trigger by modality or study type)
- 4. Execute the application automatically or manually from the viewer interface
- 5. View and export results (e.g., segmentations, reports) within their institutional PACS
All processing remains on-premise. No data is transmitted externally. Application versioning and logs ensure traceability.
Outcomes and Blueprint Implications
Lessons learned from deployment included the importance of early alignment with DICOM IOD standards and close coordination during UI integration. During the containerization phase, we encountered minor adjustments to viewer interactivity and study selection workflows. These were resolved in collaboration with platform engineers to ensure consistency across installations.
Over 10,000 institutions have access to this application since its publication. Most installations complete in under one hour, with no scripting or infrastructure changes required. Feedback highlights ease of configuration, integration into existing workflows, and the ability to conduct reproducible research at scale.
For developers, this case provides a pathway for externalizing research tools using open-source AI frameworks and enterprise-grade delivery platforms. For research users, it offers an opportunity to access robust, validated AI applications with minimal IT overhead.
Looking Ahead
Ongoing efforts include evaluating the deployed tool in multicenter research collaborations. Preliminary integration into broader research networks is underway, and future versions may support federated learning feedback, provided governance conditions allow.
Interested in Deploying Your AI Research?
Learn more about how MONAI Deploy can help scale your medical imaging AI applications across institutions worldwide.